7. Dataset loading utilities
This package also features helpers to fetch larger datasets commonlyused by the machine learning community to benchmark algorithms on datathat comes from the ‘real world’.
To evaluate the impact of the scale of the dataset (n_samples andn_features) while controlling the statistical properties of the data(typically the correlation and informativeness of the features), it isalso possible to generate synthetic data.
There are three main kinds of dataset interfaces that can be used to getdatasets depending on the desired type of dataset.
The dataset loaders. They can be used to load small standard datasets,described in the section.
The dataset fetchers. They can be used to download and load larger datasets,described in the Real world datasets section.
Both loaders and fetchers functions return a dictionary-like object holdingat least two items: an array of shape n_samples * n_features withkey data (except for 20newsgroups) and a numpy array oflength n_samples, containing the target values, with key target.
It’s also possible for almost all of these function to constrain the outputto be a tuple containing only the data and the target, by setting thereturn_X_y parameter to True.
The datasets also contain a full description in their DESCR attribute andsome contain feature_names and target_names. See the datasetdescriptions below for details.
The dataset generation functions. They can be used to generate controlledsynthetic datasets, described in the section.
These functions return a tuple (X, y) consisting of a n_samples *n_features numpy array X and an array of length n_samplescontaining the targets y.
In addition, there are also miscellaneous tools to load datasets of otherformats or from other locations, described in the Loading other datasetssection.
scikit-learn comes with a few small standard datasets that do not require todownload any file from some external website.
They can be loaded using the following functions:
These datasets are useful to quickly illustrate the behavior of thevarious algorithms implemented in scikit-learn. They are however often toosmall to be representative of real world machine learning tasks.
Data Set Characteristics:
This is a copy of UCI ML housing dataset.
This dataset was taken from the StatLib library which is maintained at Carnegie Mellon University.
The Boston house-price data of Harrison, D. and Rubinfeld, D.L. ‘Hedonicprices and the demand for clean air’, J. Environ. Economics & Management,vol.5, 81-102, 1978. Used in Belsley, Kuh & Welsch, ‘Regression diagnostics…’, Wiley, 1980. N.B. Various transformations are used in the table onpages 244-261 of the latter.
The Boston house-price data has been used in many machine learning papers that address regressionproblems.
References
Belsley, Kuh & Welsch, ‘Regression diagnostics: Identifying Influential Data and Sources of Collinearity’, Wiley, 1980. 244-261.
Quinlan,R. (1993). Combining Instance-Based and Model-Based Learning. In Proceedings on the Tenth International Conference of Machine Learning, 236-243, University of Massachusetts, Amherst. Morgan Kaufmann.
7.2.2. Iris plants dataset
Data Set Characteristics:
- Number of Instances
150 (50 in each of three classes)
- Number of Attributes
4 numeric, predictive attributes and the class
- Attribute Information
sepal length in cm
sepal width in cm
petal length in cm
petal width in cm
- class:
Iris-Setosa
Iris-Versicolour
Iris-Virginica
- Summary Statistics
sepal length:
4.3
7.9
5.84
0.83
0.7826
sepal width:
2.0
4.4
3.05
0.43
-0.4194
petal length:
1.0
6.9
3.76
1.76
0.9490 (high!)
petal width:
0.1
2.5
1.20
0.76
0.9565 (high!)
- Missing Attribute Values
None
- Class Distribution
33.3% for each of 3 classes.
- Creator
R.A. Fisher
- Donor
Michael Marshall (MARSHALL%PLU@io.arc.nasa.gov)
- Date
July, 1988
The famous Iris database, first used by Sir R.A. Fisher. The dataset is takenfrom Fisher’s paper. Note that it’s the same as in R, but not as in the UCIMachine Learning Repository, which has two wrong data points.
This is perhaps the best known database to be found in thepattern recognition literature. Fisher’s paper is a classic in the field andis referenced frequently to this day. (See Duda & Hart, for example.) Thedata set contains 3 classes of 50 instances each, where each class refers to atype of iris plant. One class is linearly separable from the other 2; thelatter are NOT linearly separable from each other.
References
Fisher, R.A. “The use of multiple measurements in taxonomic problems”Annual Eugenics, 7, Part II, 179-188 (1936); also in “Contributions toMathematical Statistics” (John Wiley, NY, 1950).
Duda, R.O., & Hart, P.E. (1973) Pattern Classification and Scene Analysis.(Q327.D83) John Wiley & Sons. ISBN 0-471-22361-1. See page 218.
Dasarathy, B.V. (1980) “Nosing Around the Neighborhood: A New SystemStructure and Classification Rule for Recognition in Partially ExposedEnvironments”. IEEE Transactions on Pattern Analysis and MachineIntelligence, Vol. PAMI-2, No. 1, 67-71.
Gates, G.W. (1972) “The Reduced Nearest Neighbor Rule”. IEEE Transactionson Information Theory, May 1972, 431-433.
See also: 1988 MLC Proceedings, 54-64. Cheeseman et al”s AUTOCLASS IIconceptual clustering system finds 3 classes in the data.
Many, many more …
7.2.3. Diabetes dataset
Ten baseline variables, age, sex, body mass index, average bloodpressure, and six blood serum measurements were obtained for each of n =442 diabetes patients, as well as the response of interest, aquantitative measure of disease progression one year after baseline.
Data Set Characteristics:
- Number of Instances
442
- Number of Attributes
First 10 columns are numeric predictive values
- Target
Column 11 is a quantitative measure of disease progression one year after baseline
- Attribute Information
Age
Sex
Body mass index
Average blood pressure
S1
S2
S3
S4
S5
S6
Note: Each of these 10 feature variables have been mean centered and scaled by the standard deviation times n_samples (i.e. the sum of squares of each column totals 1).
Source URL:
For more information see:Bradley Efron, Trevor Hastie, Iain Johnstone and Robert Tibshirani (2004) “Least Angle Regression,” Annals of Statistics (with discussion), 407-499.(https://web.stanford.edu/~hastie/Papers/LARS/LeastAngle_2002.pdf)
7.2.4. Optical recognition of handwritten digits dataset
Data Set Characteristics:
- Number of Instances
5620
- Number of Attributes
64
- Attribute Information
8x8 image of integer pixels in the range 0..16.
- Missing Attribute Values
None
- Creator
Alpaydin (alpaydin ‘@’ boun.edu.tr)
- Date
July; 1998
This is a copy of the test set of the UCI ML hand-written digits datasets
The data set contains images of hand-written digits: 10 classes whereeach class refers to a digit.
Preprocessing programs made available by NIST were used to extractnormalized bitmaps of handwritten digits from a preprinted form. From atotal of 43 people, 30 contributed to the training set and different 13to the test set. 32x32 bitmaps are divided into nonoverlapping blocks of4x4 and the number of on pixels are counted in each block. This generatesan input matrix of 8x8 where each element is an integer in the range0..16. This reduces dimensionality and gives invariance to smalldistortions.
For info on NIST preprocessing routines, see M. D. Garris, J. L. Blue, G.T. Candela, D. L. Dimmick, J. Geist, P. J. Grother, S. A. Janet, and C.L. Wilson, NIST Form-Based Handprint Recognition System, NISTIR 5469,1994.
References
C. Kaynak (1995) Methods of Combining Multiple Classifiers and TheirApplications to Handwritten Digit Recognition, MSc Thesis, Institute ofGraduate Studies in Science and Engineering, Bogazici University.
- Alpaydin, C. Kaynak (1998) Cascading Classifiers, Kybernetika.
Ken Tang and Ponnuthurai N. Suganthan and Xi Yao and A. Kai Qin.Linear dimensionalityreduction using relevance weighted LDA. School ofElectrical and Electronic Engineering Nanyang Technological University.2005.
Claudio Gentile. A New Approximate Maximal Margin ClassificationAlgorithm. NIPS. 2000.
7.2.5. Linnerrud dataset
Data Set Characteristics:
- Number of Instances
20
- Number of Attributes
3
- Missing Attribute Values
None
The Linnerud dataset constains two small dataset:
- physiological - CSV containing 20 observations on 3 exercise variables:
- Weight, Waist and Pulse.
- exercise - CSV containing 20 observations on 3 physiological variables:
- Chins, Situps and Jumps.
References
- Tenenhaus, M. (1998). La regression PLS: theorie et pratique. Paris: Editions Technic.
7.2.6. Wine recognition dataset
Data Set Characteristics:
This is a copy of UCI ML Wine recognition datasets.https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data
The data is the results of a chemical analysis of wines grown in the sameregion in Italy by three different cultivators. There are thirteen differentmeasurements taken for different constituents found in the three types ofwine.
Original Owners:
Forina, M. et al, PARVUS -An Extendible Package for Data Exploration, Classification and Correlation.Institute of Pharmaceutical and Food Analysis and Technologies,Via Brigata Salerno, 16147 Genoa, Italy.
Citation:
Lichman, M. (2013). UCI Machine Learning Repository[]. Irvine, CA: University of California,School of Information and Computer Science.
References
(1) S. Aeberhard, D. Coomans and O. de Vel,Comparison of Classifiers in High Dimensional Settings,Tech. Rep. no. 92-02, (1992), Dept. of Computer Science and Dept. ofMathematics and Statistics, James Cook University of North Queensland.(Also submitted to Technometrics).
The data was used with many others for comparing variousclassifiers. The classes are separable, though only RDAhas achieved 100% correct classification.(RDA : 100%, QDA 99.4%, LDA 98.9%, 1NN 96.1% (z-transformed data))(All results using the leave-one-out technique)
(2) S. Aeberhard, D. Coomans and O. de Vel,“THE CLASSIFICATION PERFORMANCE OF RDA”Tech. Rep. no. 92-01, (1992), Dept. of Computer Science and Dept. ofMathematics and Statistics, James Cook University of North Queensland.(Also submitted to Journal of Chemometrics).
7.2.7. Breast cancer wisconsin (diagnostic) dataset
Data Set Characteristics:
- Number of Instances
569
- Number of Attributes
30 numeric, predictive attributes and the class
- Attribute Information
radius (mean of distances from center to points on the perimeter)
texture (standard deviation of gray-scale values)
area
smoothness (local variation in radius lengths)
compactness (perimeter^2 / area - 1.0)
concavity (severity of concave portions of the contour)
concave points (number of concave portions of the contour)
symmetry
fractal dimension (“coastline approximation” - 1)
The mean, standard error, and “worst” or largest (mean of the threelargest values) of these features were computed for each image,resulting in 30 features. For instance, field 3 is Mean Radius, field13 is Radius SE, field 23 is Worst Radius.
- class:
WDBC-Malignant
WDBC-Benign
- Summary Statistics
radius (mean):
6.981
28.11
texture (mean):
9.71
39.28
perimeter (mean):
43.79
188.5
area (mean):
143.5
2501.0
smoothness (mean):
0.053
0.163
compactness (mean):
0.019
0.345
concavity (mean):
0.0
0.427
concave points (mean):
0.0
0.201
symmetry (mean):
0.106
0.304
fractal dimension (mean):
0.05
0.097
radius (standard error):
0.112
2.873
texture (standard error):
0.36
4.885
perimeter (standard error):
0.757
21.98
area (standard error):
6.802
542.2
smoothness (standard error):
0.002
0.031
compactness (standard error):
0.002
0.135
concavity (standard error):
0.0
0.396
concave points (standard error):
0.0
0.053
symmetry (standard error):
0.008
0.079
fractal dimension (standard error):
0.001
0.03
radius (worst):
7.93
36.04
texture (worst):
12.02
49.54
perimeter (worst):
50.41
251.2
area (worst):
185.2
4254.0
smoothness (worst):
0.071
0.223
compactness (worst):
0.027
1.058
concavity (worst):
0.0
1.252
concave points (worst):
0.0
0.291
symmetry (worst):
0.156
0.664
fractal dimension (worst):
0.055
0.208
- Missing Attribute Values
None
- Class Distribution
212 - Malignant, 357 - Benign
- Creator
Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian
- Donor
Nick Street
- Date
November, 1995
This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets.https://goo.gl/U2Uwz2
Features are computed from a digitized image of a fine needleaspirate (FNA) of a breast mass. They describecharacteristics of the cell nuclei present in the image.
Separating plane described above was obtained usingMultisurface Method-Tree (MSM-T) [K. P. Bennett, “Decision TreeConstruction Via Linear Programming.” Proceedings of the 4thMidwest Artificial Intelligence and Cognitive Science Society,pp. 97-101, 1992], a classification method which uses linearprogramming to construct a decision tree. Relevant featureswere selected using an exhaustive search in the space of 1-4features and 1-3 separating planes.
The actual linear program used to obtain the separating planein the 3-dimensional space is that described in:[K. P. Bennett and O. L. Mangasarian: “Robust LinearProgramming Discrimination of Two Linearly Inseparable Sets”,Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server:
ftp ftp.cs.wisc.educd math-prog/cpo-dataset/machine-learn/WDBC/
References
W.N. Street, W.H. Wolberg and O.L. Mangasarian. Nuclear feature extractionfor breast tumor diagnosis. IS&T/SPIE 1993 International Symposium onElectronic Imaging: Science and Technology, volume 1905, pages 861-870,San Jose, CA, 1993.
O.L. Mangasarian, W.N. Street and W.H. Wolberg. Breast cancer diagnosis andprognosis via linear programming. Operations Research, 43(4), pages 570-577,July-August 1995.
W.H. Wolberg, W.N. Street, and O.L. Mangasarian. Machine learning techniquesto diagnose breast cancer from fine-needle aspirates. Cancer Letters 77 (1994)163-171.
scikit-learn provides tools to load larger datasets, downloading them ifnecessary.
They can be loaded using the following functions:
| ([data_home, shuffle, …]) | Load the Olivetti faces data-set from AT&T (classification). |
fetch_20newsgroups([data_home, subset, …]) | Load the filenames and data from the 20 newsgroups dataset (classification). |
| ([subset, …]) | Load the 20 newsgroups dataset and vectorize it into token counts (classification). |
fetch_lfw_people([data_home, funneled, …]) | Load the Labeled Faces in the Wild (LFW) people dataset (classification). |
| ([subset, data_home, …]) | Load the Labeled Faces in the Wild (LFW) pairs dataset (classification). |
fetch_covtype([data_home, …]) | Load the covertype dataset (classification). |
| ([data_home, subset, …]) | Load the RCV1 multilabel dataset (classification). |
fetch_kddcup99([subset, data_home, shuffle, …]) | Load the kddcup99 dataset (classification). |
| ([data_home, …]) | Load the California housing dataset (regression). |
This dataset contains a set of face images taken between April 1992 andApril 1994 at AT&T Laboratories Cambridge. The function is the datafetching / caching function that downloads the dataarchive from AT&T.
As described on the original website:
There are ten different images of each of 40 distinct subjects. For somesubjects, the images were taken at different times, varying the lighting,facial expressions (open / closed eyes, smiling / not smiling) and facialdetails (glasses / no glasses). All the images were taken against a darkhomogeneous background with the subjects in an upright, frontal position(with tolerance for some side movement).
Data Set Characteristics:
Classes
40
Samples total
400
Dimensionality
4096
Features
real, between 0 and 1
The image is quantized to 256 grey levels and stored as unsigned 8-bitintegers; the loader will convert these to floating point values on theinterval [0, 1], which are easier to work with for many algorithms.
The “target” for this database is an integer from 0 to 39 indicating theidentity of the person pictured; however, with only 10 examples per class, thisrelatively small dataset is more interesting from an unsupervised orsemi-supervised perspective.
The original dataset consisted of 92 x 112, while the version available hereconsists of 64x64 images.
When using these images, please give credit to AT&T Laboratories Cambridge.
7.3.2. The 20 newsgroups text dataset
The 20 newsgroups dataset comprises around 18000 newsgroups posts on20 topics split in two subsets: one for training (or development)and the other one for testing (or for performance evaluation). The splitbetween the train and test set is based upon a messages posted beforeand after a specific date.
This module contains two loaders. The first one,sklearn.datasets.fetch_20newsgroups,returns a list of the raw texts that can be fed to text featureextractors such as with custom parameters so as to extract feature vectors.The second one, sklearn.datasets.fetch_20newsgroups_vectorized,returns ready-to-use features, i.e., it is not necessary to use a featureextractor.
Data Set Characteristics:
7.3.2.1. Usage
The function is a datafetching / caching functions that downloads the data archive fromthe original 20 newsgroups website, extracts the archive contentsin the ~/scikit_learn_data/20news_home folder and calls the on either the training ortesting set folder, or both of them:
>>>
The real data lies in the and target attributes. The targetattribute is the integer index of the category:
>>>
- >>> newsgroups_train.filenames.shape
- (11314,)
- >>> newsgroups_train.target.shape
- (11314,)
- >>> newsgroups_train.target[:10]
- array([ 7, 4, 4, 1, 14, 16, 13, 3, 2, 4])
It is possible to load only a sub-selection of the categories by passing thelist of the categories to load to thesklearn.datasets.fetch_20newsgroups function:
>>>
- >>> cats = ['alt.atheism', 'sci.space']
- >>> newsgroups_train = fetch_20newsgroups(subset='train', categories=cats)
- >>> list(newsgroups_train.target_names)
- ['alt.atheism', 'sci.space']
- >>> newsgroups_train.filenames.shape
- (1073,)
- >>> newsgroups_train.target.shape
- (1073,)
- >>> newsgroups_train.target[:10]
- array([0, 1, 1, 1, 0, 1, 1, 0, 0, 0])
7.3.2.2. Converting text to vectors
In order to feed predictive or clustering models with the text data,one first need to turn the text into vectors of numerical values suitablefor statistical analysis. This can be achieved with the utilities of thesklearn.feature_extraction.text as demonstrated in the followingexample that extract vectors of unigram tokensfrom a subset of 20news:
>>>
- >>> from sklearn.feature_extraction.text import TfidfVectorizer
- >>> categories = ['alt.atheism', 'talk.religion.misc',
- ... 'comp.graphics', 'sci.space']
- >>> newsgroups_train = fetch_20newsgroups(subset='train',
- ... categories=categories)
- >>> vectorizer = TfidfVectorizer()
- >>> vectors = vectorizer.fit_transform(newsgroups_train.data)
- >>> vectors.shape
- (2034, 34118)
The extracted TF-IDF vectors are very sparse, with an average of 159 non-zerocomponents by sample in a more than 30000-dimensional space(less than .5% non-zero features):
>>>
- >>> vectors.nnz / float(vectors.shape[0])
- 159.01327...
sklearn.datasets.fetch_20newsgroups_vectorized is a function whichreturns ready-to-use token counts features instead of file names.
7.3.2.3. Filtering text for more realistic training
It is easy for a classifier to overfit on particular things that appear in the20 Newsgroups data, such as newsgroup headers. Many classifiers achieve veryhigh F-scores, but their results would not generalize to other documents thataren’t from this window of time.
For example, let’s look at the results of a multinomial Naive Bayes classifier,which is fast to train and achieves a decent F-score:
>>>
- >>> from sklearn.naive_bayes import MultinomialNB
- >>> from sklearn import metrics
- >>> newsgroups_test = fetch_20newsgroups(subset='test',
- ... categories=categories)
- >>> vectors_test = vectorizer.transform(newsgroups_test.data)
- >>> clf = MultinomialNB(alpha=.01)
- >>> clf.fit(vectors, newsgroups_train.target)
- MultinomialNB(alpha=0.01, class_prior=None, fit_prior=True)
- >>> pred = clf.predict(vectors_test)
- >>> metrics.f1_score(newsgroups_test.target, pred, average='macro')
- 0.88213...
(The example shufflesthe training and test data, instead of segmenting by time, and in that casemultinomial Naive Bayes gets a much higher F-score of 0.88. Are you suspiciousyet of what’s going on inside this classifier?)
Let’s take a look at what the most informative features are:
>>>
- >>> import numpy as np
- >>> def show_top10(classifier, vectorizer, categories):
- ... feature_names = np.asarray(vectorizer.get_feature_names())
- ... for i, category in enumerate(categories):
- ... top10 = np.argsort(classifier.coef_[i])[-10:]
- ... print("%s: %s" % (category, " ".join(feature_names[top10])))
- ...
- >>> show_top10(clf, vectorizer, newsgroups_train.target_names)
- alt.atheism: edu it and in you that is of to the
- comp.graphics: edu in graphics it is for and of to the
- sci.space: edu it that is in and space to of the
- talk.religion.misc: not it you in is that and to of the
You can now see many things that these features have overfit to:
Almost every group is distinguished by whether headers such as
NNTP-Posting-Host:andDistribution:appear more or less often.Another significant feature involves whether the sender is affiliated witha university, as indicated either by their headers or their signature.
The word “article” is a significant feature, based on how often people quoteprevious posts like this: “In article [article ID], [name] <[e-mail address]>wrote:”
Other features match the names and e-mail addresses of particular people whowere posting at the time.
For this reason, the functions that load 20 Newsgroups data provide aparameter called remove, telling it what kinds of information to strip outof each file. remove should be a tuple containing any subset of('headers', 'footers', 'quotes'), telling it to remove headers, signatureblocks, and quotation blocks respectively.
>>>
- >>> newsgroups_test = fetch_20newsgroups(subset='test',
- ... remove=('headers', 'footers', 'quotes'),
- ... categories=categories)
- >>> vectors_test = vectorizer.transform(newsgroups_test.data)
- >>> pred = clf.predict(vectors_test)
- >>> metrics.f1_score(pred, newsgroups_test.target, average='macro')
- 0.77310...
This classifier lost over a lot of its F-score, just because we removedmetadata that has little to do with topic classification.It loses even more if we also strip this metadata from the training data:
>>>
- >>> newsgroups_train = fetch_20newsgroups(subset='train',
- ... remove=('headers', 'footers', 'quotes'),
- ... categories=categories)
- >>> vectors = vectorizer.fit_transform(newsgroups_train.data)
- >>> clf = MultinomialNB(alpha=.01)
- >>> clf.fit(vectors, newsgroups_train.target)
- MultinomialNB(alpha=0.01, class_prior=None, fit_prior=True)
>>>
- >>> vectors_test = vectorizer.transform(newsgroups_test.data)
- >>> pred = clf.predict(vectors_test)
- >>> metrics.f1_score(newsgroups_test.target, pred, average='macro')
- 0.76995...
Some other classifiers cope better with this harder version of the task. Tryrunning Sample pipeline for text feature extraction and evaluation with and withoutthe —filter option to compare the results.
Recommendation
When evaluating text classifiers on the 20 Newsgroups data, youshould strip newsgroup-related metadata. In scikit-learn, you can do this bysetting remove=('headers', 'footers', 'quotes'). The F-score will belower because it is more realistic.
Examples
7.3.3. The Labeled Faces in the Wild face recognition dataset
This dataset is a collection of JPEG pictures of famous people collectedover the internet, all details are available on the official website:
Each picture is centered on a single face. The typical task is calledFace Verification: given a pair of two pictures, a binary classifiermust predict whether the two images are from the same person.
An alternative task, Face Recognition or Face Identification is:given the picture of the face of an unknown person, identify the nameof the person by referring to a gallery of previously seen pictures ofidentified persons.
Both Face Verification and Face Recognition are tasks that are typicallyperformed on the output of a model trained to perform Face Detection. Themost popular model for Face Detection is called Viola-Jones and isimplemented in the OpenCV library. The LFW faces were extracted by thisface detector from various online websites.
Data Set Characteristics:
Classes
5749
Samples total
13233
Dimensionality
5828
Features
real, between 0 and 255
7.3.3.1. Usage
scikit-learn provides two loaders that will automatically download,cache, parse the metadata files, decode the jpeg and convert theinteresting slices into memmapped numpy arrays. This dataset size is morethan 200 MB. The first load typically takes more than a couple of minutesto fully decode the relevant part of the JPEG files into numpy arrays. Ifthe dataset has been loaded once, the following times the loading timesless than 200ms by using a memmapped version memoized on the disk in the~/scikit_learn_data/lfw_home/ folder using joblib.
The first loader is used for the Face Identification task: a multi-classclassification task (hence supervised learning):
>>>
The default slice is a rectangular shape around the face, removingmost of the background:
>>>
- >>> lfw_people.data.dtype
- dtype('float32')
- >>> lfw_people.data.shape
- (1288, 1850)
- >>> lfw_people.images.shape
- (1288, 50, 37)
Each of the 1140 faces is assigned to a single person id in the targetarray:
>>>
- >>> lfw_people.target.shape
- (1288,)
- >>> list(lfw_people.target[:10])
- [5, 6, 3, 1, 0, 1, 3, 4, 3, 0]
The second loader is typically used for the face verification task: each sampleis a pair of two picture belonging or not to the same person:
>>>
- >>> from sklearn.datasets import fetch_lfw_pairs
- >>> lfw_pairs_train = fetch_lfw_pairs(subset='train')
- >>> list(lfw_pairs_train.target_names)
- ['Different persons', 'Same person']
- (2200, 2, 62, 47)
- >>> lfw_pairs_train.data.shape
- (2200, 5828)
- >>> lfw_pairs_train.target.shape
- (2200,)
Both for the andsklearn.datasets.fetch_lfw_pairs function it ispossible to get an additional dimension with the RGB color channels bypassing color=True, in that case the shape will be(2200, 2, 62, 47, 3).
The datasets is subdivided into3 subsets: the development train set, the development test set andan evaluation 10_folds set meant to compute performance metrics using a10-folds cross validation scheme.
References:
- Labeled Faces in the Wild: A Database for Studying Face Recognitionin Unconstrained Environments.Gary B. Huang, Manu Ramesh, Tamara Berg, and Erik Learned-Miller.University of Massachusetts, Amherst, Technical Report 07-49, October, 2007.
7.3.3.2. Examples
7.3.4. Forest covertypes
The samples in this dataset correspond to 30×30m patches of forest in the US,collected for the task of predicting each patch’s cover type,i.e. the dominant species of tree.There are seven covertypes, making this a multiclass classification problem.Each sample has 54 features, described on thedataset’s homepage.Some of the features are boolean indicators,while others are discrete or continuous measurements.
Data Set Characteristics:
Classes
7
Samples total
581012
Dimensionality
54
Features
int
will load the covertype dataset;it returns a dictionary-like objectwith the feature matrix in the data memberand the target values in target.The dataset will be downloaded from the web if necessary.
7.3.5. RCV1 dataset
Reuters Corpus Volume I (RCV1) is an archive of over 800,000 manuallycategorized newswire stories made available by Reuters, Ltd. for researchpurposes. The dataset is extensively described in 1.
Data Set Characteristics:
Classes
103
Samples total
804414
Dimensionality
47236
Features
real, between 0 and 1
will load the followingversion: RCV1-v2, vectors, full sets, topics multilabels:
>>>
- >>> from sklearn.datasets import fetch_rcv1
- >>> rcv1 = fetch_rcv1()
It returns a dictionary-like object, with the following attributes:
data:The feature matrix is a scipy CSR sparse matrix, with 804414 samples and47236 features. Non-zero values contains cosine-normalized, log TF-IDF vectors.A nearly chronological split is proposed in 1: The first 23149 samples arethe training set. The last 781265 samples are the testing set. This followsthe official LYRL2004 chronological split. The array has 0.16% of non zerovalues:
>>>
- >>> rcv1.data.shape
- (804414, 47236)
target:The target values are stored in a scipy CSR sparse matrix, with 804414 samplesand 103 categories. Each sample has a value of 1 in its categories, and 0 inothers. The array has 3.15% of non zero values:
>>>
- >>> rcv1.target.shape
- (804414, 103)
sample_id:Each sample can be identified by its ID, ranging (with gaps) from 2286to 810596:
>>>
- >>> rcv1.sample_id[:3]
- array([2286, 2287, 2288], dtype=uint32)
target_names:The target values are the topics of each sample. Each sample belongs to atleast one topic, and to up to 17 topics. There are 103 topics, eachrepresented by a string. Their corpus frequencies span five orders ofmagnitude, from 5 occurrences for ‘GMIL’, to 381327 for ‘CCAT’:
>>>
- >>> rcv1.target_names[:3].tolist()
- ['E11', 'ECAT', 'M11']
The dataset will be downloaded from the if necessary.The compressed size is about 656 MB.
References
- 1(1,)
- Lewis, D. D., Yang, Y., Rose, T. G., & Li, F. (2004).RCV1: A new benchmark collection for text categorization research.The Journal of Machine Learning Research, 5, 361-397.
7.3.6. Kddcup 99 dataset
The KDD Cup ‘99 dataset was created by processing the tcpdump portionsof the 1998 DARPA Intrusion Detection System (IDS) Evaluation dataset,created by MIT Lincoln Lab [1]. The artificial data (described on the dataset’shomepage) wasgenerated using a closed network and hand-injected attacks to produce alarge number of different types of attack with normal activity in thebackground. As the initial goal was to produce a large training set forsupervised learning algorithms, there is a large proportion (80.1%) ofabnormal data which is unrealistic in real world, and inappropriate forunsupervised anomaly detection which aims at detecting ‘abnormal’ data, ie
qualitatively different from normal data
in large minority among the observations.
We thus transform the KDD Data set into two different data sets: SA and SF.
-SA is obtained by simply selecting all the normal data, and a smallproportion of abnormal data to gives an anomaly proportion of 1%.
-SF is obtained as in [2]by simply picking up the data whose attribute logged_in is positive, thusfocusing on the intrusion attack, which gives a proportion of 0.3% ofattack.
-http and smtp are two subsets of SF corresponding with third featureequal to ‘http’ (resp. to ‘smtp’)
General KDD structure :
Samples total
4898431
Dimensionality
41
Features
discrete (int) or continuous (float)
Targets
str, ‘normal.’ or name of the anomaly type
SA structure :
Samples total
976158
Dimensionality
41
Features
discrete (int) or continuous (float)
Targets
str, ‘normal.’ or name of the anomaly type
SF structure :
http structure :
Samples total
619052
Dimensionality
3
Features
discrete (int) or continuous (float)
Targets
str, ‘normal.’ or name of the anomaly type
smtp structure :
Samples total
95373
Dimensionality
3
Features
discrete (int) or continuous (float)
Targets
str, ‘normal.’ or name of the anomaly type
will load the kddcup99 dataset; itreturns a dictionary-like object with the feature matrix in the data memberand the target values in target. The dataset will be downloaded from theweb if necessary.
7.3.7. California Housing dataset
Data Set Characteristics:
This dataset was obtained from the StatLib repository.http://lib.stat.cmu.edu/datasets/
The target variable is the median house value for California districts.
This dataset was derived from the 1990 U.S. census, using one row per censusblock group. A block group is the smallest geographical unit for which the U.S.Census Bureau publishes sample data (a block group typically has a populationof 600 to 3,000 people).
It can be downloaded/loaded using the function.
References
- Pace, R. Kelley and Ronald Barry, Sparse Spatial Autoregressions,Statistics and Probability Letters, 33 (1997) 291-297
In addition, scikit-learn includes various random sample generators thatcan be used to build artificial datasets of controlled size and complexity.
These generators produce a matrix of features and corresponding discretetargets.
7.4.1.1. Single label
Both make_blobs and create multiclassdatasets by allocating each class one or more normally-distributed clusters ofpoints. make_blobs provides greater control regarding the centers andstandard deviations of each cluster, and is used to demonstrate clustering. specialises in introducing noise by way of:correlated, redundant and uninformative features; multiple Gaussian clustersper class; and linear transformations of the feature space.
make_gaussian_quantiles divides a single Gaussian cluster intonear-equal-size classes separated by concentric hyperspheres. generates a similar binary, 10-dimensional problem. and make_moons generate 2d binary classificationdatasets that are challenging to certain algorithms (e.g. centroid-basedclustering or linear classification), including optional Gaussian noise.They are useful for visualisation. produces Gaussian datawith a spherical decision boundary for binary classification, whilemake_moons produces two interleaving half circles.
7.4.1.2. Multilabel
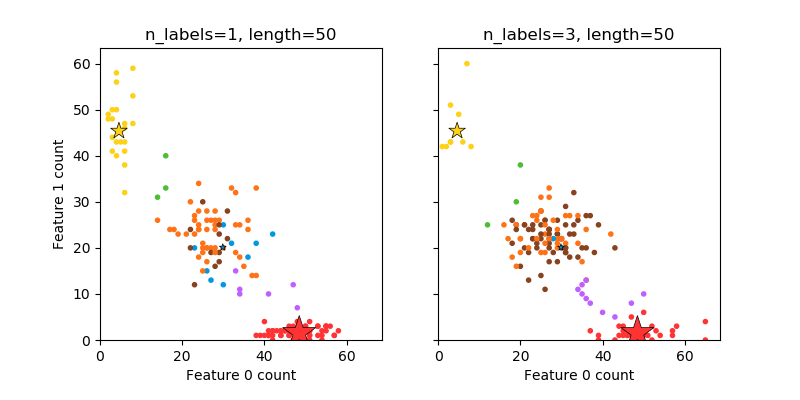
generates random samples with multiplelabels, reflecting a bag of words drawn from a mixture of topics. The number oftopics for each document is drawn from a Poisson distribution, and the topicsthemselves are drawn from a fixed random distribution. Similarly, the number ofwords is drawn from Poisson, with words drawn from a multinomial, where eachtopic defines a probability distribution over words. Simplifications withrespect to true bag-of-words mixtures include:
Per-topic word distributions are independently drawn, where in reality allwould be affected by a sparse base distribution, and would be correlated.
For a document generated from multiple topics, all topics are weightedequally in generating its bag of words.
Documents without labels words at random, rather than from a basedistribution.
7.4.1.3. Biclustering
| (shape, n_clusters[, noise, …]) | Generate an array with constant block diagonal structure for biclustering. |
make_checkerboard(shape, n_clusters[, …]) | Generate an array with block checkerboard structure for biclustering. |
7.4.2. Generators for regression
produces regression targets as an optionally-sparserandom linear combination of random features, with noise. Its informativefeatures may be uncorrelated, or low rank (few features account for most of thevariance).
Other regression generators generate functions deterministically fromrandomized features. make_sparse_uncorrelated produces a target as alinear combination of four features with fixed coefficients.Others encode explicitly non-linear relations: is related by polynomial and sine transforms;make_friedman2 includes feature multiplication and reciprocation; and is similar with an arctan transformation on the target.
7.4.3. Generators for manifold learning
make_s_curve([n_samples, noise, random_state]) | Generate an S curve dataset. |
| ([n_samples, noise, random_state]) | Generate a swiss roll dataset. |
7.4.4. Generators for decomposition
make_low_rank_matrix([n_samples, …]) | Generate a mostly low rank matrix with bell-shaped singular values |
| (n_samples, …[, …]) | Generate a signal as a sparse combination of dictionary elements. |
make_spd_matrix(n_dim[, random_state]) | Generate a random symmetric, positive-definite matrix. |
| ([dim, alpha, …]) | Generate a sparse symmetric definite positive matrix. |
7.5.1. Sample images
Scikit-learn also embed a couple of sample JPEG images published under CreativeCommons license by their authors. Those images can be useful to test algorithmsand pipeline on 2D data.
Warning
The default coding of images is based on the uint8 dtype tospare memory. Often machine learning algorithms work best if theinput is converted to a floating point representation first. Also,if you plan to use matplotlib.pyplpt.imshow don’t forget to scale to the range0 - 1 as done in the following example.
Examples:
7.5.2. Datasets in svmlight / libsvm format
scikit-learn includes utility functions for loadingdatasets in the svmlight / libsvm format. In this format, each linetakes the form <label> <feature-id>:<feature-value><feature-id>:<feature-value> …. This format is especially suitable for sparse datasets.In this module, scipy sparse CSR matrices are used for X and numpy arrays are used for y.
You may load a dataset like as follows:
>>>
- >>> from sklearn.datasets import load_svmlight_file
- >>> X_train, y_train = load_svmlight_file("/path/to/train_dataset.txt")
- ...
You may also load two (or more) datasets at once:
>>>
In this case, X_train and X_test are guaranteed to have the same numberof features. Another way to achieve the same result is to fix the number offeatures:
>>>
- >>> X_test, y_test = load_svmlight_file(
- ... "/path/to/test_dataset.txt", n_features=X_train.shape[1])
- ...
Related links:
Public datasets in svmlight / libsvm format: https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets
Faster API-compatible implementation:
7.5.3. Downloading datasets from the openml.org repository
openml.org is a public repository for machine learningdata and experiments, that allows everybody to upload open datasets.
The sklearn.datasets package is able to download datasetsfrom the repository using the function.
For example, to download a dataset of gene expressions in mice brains:
>>>
- >>> from sklearn.datasets import fetch_openml
- >>> mice = fetch_openml(name='miceprotein', version=4)
To fully specify a dataset, you need to provide a name and a version, thoughthe version is optional, see Dataset Versions below.The dataset contains a total of 1080 examples belonging to 8 differentclasses:
>>>
- >>> mice.data.shape
- (1080, 77)
- >>> mice.target.shape
- (1080,)
- >>> np.unique(mice.target)
- array(['c-CS-m', 'c-CS-s', 'c-SC-m', 'c-SC-s', 't-CS-m', 't-CS-s', 't-SC-m', 't-SC-s'], dtype=object)
You can get more information on the dataset by looking at the DESCRand details attributes:
>>>
- >>> print(mice.DESCR)
- **Author**: Clara Higuera, Katheleen J. Gardiner, Krzysztof J. Cios
- **Source**: [UCI](https://archive.ics.uci.edu/ml/datasets/Mice+Protein+Expression) - 2015
- **Please cite**: Higuera C, Gardiner KJ, Cios KJ (2015) Self-Organizing
- Feature Maps Identify Proteins Critical to Learning in a Mouse Model of Down
- Syndrome. PLoS ONE 10(6): e0129126...
- >>> mice.details
- {'id': '40966', 'name': 'MiceProtein', 'version': '4', 'format': 'ARFF',
- 'upload_date': '2017-11-08T16:00:15', 'licence': 'Public',
- 'url': 'https://www.openml.org/data/v1/download/17928620/MiceProtein.arff',
- 'file_id': '17928620', 'default_target_attribute': 'class',
- 'row_id_attribute': 'MouseID',
- 'ignore_attribute': ['Genotype', 'Treatment', 'Behavior'],
- 'tag': ['OpenML-CC18', 'study_135', 'study_98', 'study_99'],
- 'visibility': 'public', 'status': 'active',
- 'md5_checksum': '3c479a6885bfa0438971388283a1ce32'}
The DESCR contains a free-text description of the data, while detailscontains a dictionary of meta-data stored by openml, like the dataset id.For more details, see the The data_id of the mice protein datasetis 40966, and you can use this (or the name) to get more information on thedataset on the openml website:
>>>
- >>> mice.url
- 'https://www.openml.org/d/40966'
The data_id also uniquely identifies a dataset from OpenML:
>>>
- >>> mice = fetch_openml(data_id=40966)
- >>> mice.details
- {'id': '4550', 'name': 'MiceProtein', 'version': '1', 'format': 'ARFF',
- 'creator': ...,
- 'upload_date': '2016-02-17T14:32:49', 'licence': 'Public', 'url':
- 'https://www.openml.org/data/v1/download/1804243/MiceProtein.ARFF', 'file_id':
- '1804243', 'default_target_attribute': 'class', 'citation': 'Higuera C,
- Gardiner KJ, Cios KJ (2015) Self-Organizing Feature Maps Identify Proteins
- Critical to Learning in a Mouse Model of Down Syndrome. PLoS ONE 10(6):
- e0129126. [Web Link] journal.pone.0129126', 'tag': ['OpenML100', 'study_14',
- 'study_34'], 'visibility': 'public', 'status': 'active', 'md5_checksum':
- '3c479a6885bfa0438971388283a1ce32'}
7.5.3.1. Dataset Versions
A dataset is uniquely specified by its data_id, but not necessarily by itsname. Several different “versions” of a dataset with the same name can existwhich can contain entirely different datasets.If a particular version of a dataset has been found to contain significantissues, it might be deactivated. Using a name to specify a dataset will yieldthe earliest version of a dataset that is still active. That means thatfetch_openml(name="miceprotein") can yield different results at differenttimes if earlier versions become inactive.You can see that the dataset with data_id 40966 that we fetched above isthe version 1 of the “miceprotein” dataset:
>>>
- >>> mice.details['version']
- '1'
In fact, this dataset only has one version. The iris dataset on the other handhas multiple versions:
>>>
- >>> iris = fetch_openml(name="iris")
- >>> iris.details['version']
- '1'
- >>> iris.details['id']
- '61'
- >>> iris_61 = fetch_openml(data_id=61)
- >>> iris_61.details['version']
- '1'
- >>> iris_61.details['id']
- '61'
- >>> iris_969 = fetch_openml(data_id=969)
- >>> iris_969.details['version']
- '3'
- >>> iris_969.details['id']
- '969'
Specifying the dataset by the name “iris” yields the lowest version, version 1,with the data_id 61. To make sure you always get this exact dataset, it issafest to specify it by the dataset data_id. The other dataset, withdata_id 969, is version 3 (version 2 has become inactive), and contains abinarized version of the data:
>>>
- >>> np.unique(iris_969.target)
- array(['N', 'P'], dtype=object)
You can also specify both the name and the version, which also uniquelyidentifies the dataset:
>>>
References:
- Vanschoren, van Rijn, Bischl and Torgo“OpenML: networked science in machine learning”,ACM SIGKDD Explorations Newsletter, 15(2), 49-60, 2014.
scikit-learn works on any numeric data stored as numpy arrays or scipy sparsematrices. Other types that are convertible to numeric arrays such as pandasDataFrame are also acceptable.
Here are some recommended ways to load standard columnar data into aformat usable by scikit-learn:
provides tools to read data from common formats including CSV, Excel, JSONand SQL. DataFrames may also be constructed from lists of tuples or dicts.Pandas handles heterogeneous data smoothly and provides tools formanipulation and conversion into a numeric array suitable for scikit-learn.
scipy.iospecializes in binary formats often used in scientific computingcontext such as .mat and .arff
for standard loading of columnar data into numpy arrays
scikit-learn’s
datasets.load_svmlight_filefor the svmlight or libSVMsparse formatscikit-learn’s
datasets.load_filesfor directories of text files wherethe name of each directory is the name of each category and each file insideof each directory corresponds to one sample from that category
For some miscellaneous data such as images, videos, and audio, you may wish torefer to:
skimage.io orfor loading images and videos into numpy arrays
Categorical (or nominal) features stored as strings (common in pandas DataFrames)will need converting to numerical features using sklearn.preprocessing.OneHotEncoderor or similar.See Preprocessing data.